Applications/CellMigrationAnalyzer: Difference between revisions
From MiToBo
Jump to navigationJump to search
| (2 intermediate revisions by the same user not shown) | |||
| Line 7: | Line 7: | ||
'''Description:'''<br/> | '''Description:'''<br/> | ||
* Segments and tracks fluorescently labeled cells from time lapse image sequences | * Segments and tracks fluorescently labeled cells from time lapse image sequences | ||
* Descriptors characterizing migratory behaviour, morphological parameters and fluorescence distributions are reported | * An additional fluorescence channel acting as a marker for including/ excluding certain cells can be used | ||
* Descriptors characterizing migratory behaviour, morphological parameters and fluorescence distributions are reported in tab separated files that can be further processed by spreadsheet programs or R. | |||
'''Name of Plugin/Operator:'''<br/> | '''Name of Plugin/Operator:'''<br/> | ||
| Line 15: | Line 16: | ||
<br> | <br> | ||
(since MiToBo version 1.3) | (since MiToBo version 1.3) | ||
'''Additional operators contained in the package:'''<br/> | |||
* FluorescentCellSegmenter | |||
** segments fluorescently labeled cells from images/ image sequences | |||
* CellSegmentationPostprocessing | |||
** postprocesses objects from binary images/ image sequences (e.g. size filtering) | |||
* CellTrackerBipartite | |||
** tracks (i.e. creates a label image sequence) objects from binary image sequences based on the objects' locations and areas | |||
* MigrationAnalyzer | |||
** calculates migration and shape descriptors of objects from a label image sequence as well as intensity descriptors if a corresponding fluorescence image sequence is given | |||
** can create colored trajectories on a separate image or as overlay | |||
'''Example images'''<br/> | '''Example images'''<br/> | ||
[http://www.informatik.uni-halle.de/mitobo/downloads/tracking_examples.zip tracking sequences] | [http://www.informatik.uni-halle.de/mitobo/downloads/tracking_examples.zip tracking sequences] | ||
Latest revision as of 08:40, 3 September 2014
Cell Migration Analyzer
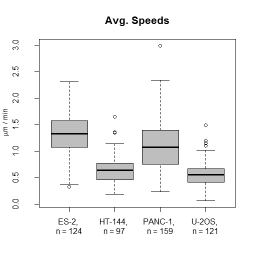
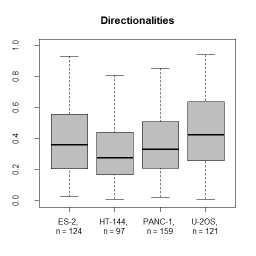
Description:
- Segments and tracks fluorescently labeled cells from time lapse image sequences
- An additional fluorescence channel acting as a marker for including/ excluding certain cells can be used
- Descriptors characterizing migratory behaviour, morphological parameters and fluorescence distributions are reported in tab separated files that can be further processed by spreadsheet programs or R.
Name of Plugin/Operator:
de.unihalle.informatik.MiToBo.apps.singleCellTracking2D.CellMigrationAnalyzer
(since MiToBo version 1.3)
Additional operators contained in the package:
- FluorescentCellSegmenter
- segments fluorescently labeled cells from images/ image sequences
- CellSegmentationPostprocessing
- postprocesses objects from binary images/ image sequences (e.g. size filtering)
- CellTrackerBipartite
- tracks (i.e. creates a label image sequence) objects from binary image sequences based on the objects' locations and areas
- MigrationAnalyzer
- calculates migration and shape descriptors of objects from a label image sequence as well as intensity descriptors if a corresponding fluorescence image sequence is given
- can create colored trajectories on a separate image or as overlay
Example images
tracking sequences