Operators/Features/MorphologyAnalyzer2D: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
The MorphologyAnalyer2D operator implements a large collection of features to characterize the shape of image regions. | The MorphologyAnalyer2D operator implements a large collection of features to characterize the shape of image regions. | ||
[[File: | [[File:MTBOperatorControlFrame-MorphologyAnalyzer2D.png|450px|right|link=]] | ||
==== Name of Plugin/Operator ==== | ==== Name of Plugin/Operator ==== | ||
| Line 10: | Line 10: | ||
==== Latest News ==== | ==== Latest News ==== | ||
The MorphologyAnalyzer2D is currently under active development. Some of the features already documented below will become available in the next version 1.8.6 of MiToBo which is to be released soon. | The MorphologyAnalyzer2D is currently under active development. Some of the features already documented below will become available<br> in the next version 1.8.6 of MiToBo which is to be released soon. | ||
==== Main features ==== | ==== Main features ==== | ||
Latest revision as of 08:40, 1 June 2017
The MorphologyAnalyer2D operator implements a large collection of features to characterize the shape of image regions.
Name of Plugin/Operator
de.unihalle.informatik.MiToBo.features.MorphologyAnalyzer2D
Latest News
The MorphologyAnalyzer2D is currently under active development. Some of the features already documented below will become available
in the next version 1.8.6 of MiToBo which is to be released soon.
Main features
- extracts a large collection of features to characterize region shape and morphology
- handles different types of input, i.e., label images and ImageJ ROIs
- optionally provides visualization of extracted features
Usage
To run the MorphologyAnalyzer2D perform the following steps:
- install MiToBo by following the instructions on the Installation page
- run MiToBo and start the operator runner by selecting the menu item MiToBo Runner from Plugins -> MiToBo
- in the selection menu navigate to 'de.unihalle.informatik.MiToBo.features' and select the operator MorphologyAnalyzer2D
This will bring up the operator window of the MorphologyAnalyzer2D.
Input data
The operator supports two different types of input data:
- label image: an image where for each region the pixels are expected to be uniquely labeled, i.e. all have the same intensity value; all pixels being labeled with 0 are treated as background
- regions (in ImageJ ROI format): here regions can directly be imported from ImageJ's ROI manager or be read from ROI files in specific formats from disk; also manual input of region data is supported
Calibration information
Many of the features refer to measuring lengths or areas of region entities. The units to be used for these measurements can freely be configured.
If appropriate calibration data is available from the given input data, e.g., as meta information in a label image, this data is automatically used and displayed.
But, the user can overwrite the calibration by manually specifiying the size of a pixel in the desired units.
E.g., with regard to microscopy images a common unit are microns and a proper calibration requires to provide information on the size of a pixel in microns.
Important note:
The operator always assumes square pixels, i.e. the pixel lengths in x and y dimension are assumed to be equal.
If this does not hold for an image or ROI data some of the feature values will be incorrect!
Lengths are first measured in pixels by the operator and areas in square pixels. Subsequently the numbers are then converted to the specified units, e.g., converted to microns or square microns based on the given size of a pixel.
Configuration of features to extract
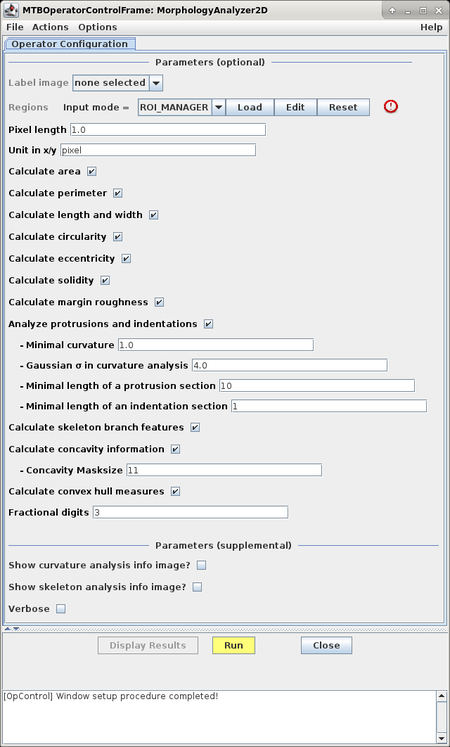
The configuration window allows to select and configure the set of features to be extracted:
| Name of Option | Description |
| Calculate area | Activates area feature <math>\scriptstyle A(R)</math> which is measured in terms of the number of pixels in a region. |
| Calculate perimeter | Activates perimeter feature <math>\scriptstyle P(R)</math> which is calculated from the 8-neighborhood contour of the region; the contour is extracted and then traced pixel by pixel where each horizontal or vertical step yields a distance of 1 and each diagonal step a distance of <math>\scriptstyle \sqrt{2}</math>; the result for the region perimeter is given by the sum of all distances between pairs of neighboring pixels along the contour. |
| Calculate length and width | Activates calculation of length <math>\scriptstyle L(R)</math> and width <math>\scriptstyle W(R)</math> of a region. The length and width of a region can be interpreted as twice the lengths of the major and minor axes of an ellipse fit to the region pixels. They are calculated as follows: <math>\scriptstyle L(R) = 2 \cdot \sqrt{\frac{2\cdot \left( \mu_{2,0} + \mu_{0,2} + \sqrt{ (\mu_{2,0}-\mu_{0,2})^2+4\mu_{1,1}^2}\right)}{\mu_{0,0}}}</math> and <math>\scriptstyle W(R) = 2 \cdot \sqrt{\frac{2\cdot \left( \mu_{2,0} + \mu_{0,2} - \sqrt{(\mu_{2,0}-\mu_{0,2})^2+4\mu_{1,1}^2}\right)}{\mu_{0,0}}}</math> The <math>\scriptstyle \mu_{k,l}</math> are central moments, <math>\scriptstyle \mu_{k,l} = \sum_{(x,y) \in R} (x- \bar{x})^k (y-\bar{y})^l</math>, with <math>\scriptstyle (\bar{x},\bar{y})</math> being the centroid of the region. For further details refer to R. Prokop/A. Reeves, A Survey of Moment-Based Techniques for Unoccluded Object Representation and Recognition, Graphical Models and Image Processing (CVGIP), pp. 438-460, 1992. |
| Calculate circularity | Activates the circularity feature <math>\scriptstyle C(R)</math> which is calculated from region area and perimeter as follows: <math>\scriptstyle C(R) = \frac{4 \pi A(R)}{P(R)^2}</math>. |
| Calculate eccentricity | Activates the eccentricity feature <math>\scriptstyle E(R)</math> which is calculated based on region moments as follows: <math>\scriptstyle E(R) = \frac{(\mu_{2,0}-\mu_{0,2})^2+4\mu_{1,1}^2}{\mu_{2,0} + \mu_{0,2}}</math>. |
| Calculate solidity | Activates the solidity feature which is defined as <math>\scriptstyle S(R) = \frac{A(R)}{A_{CH}(R)}</math> where <math>\scriptstyle A_{CH}</math> is the area of the convex hull of the region. |
| Calculate margin roughness | Activates the margin roughness feature which quantifies the deviation of the average curvature of the region contour compared to a perfect circle.
For further details refer to T. McLellan, J. Endler, The Relative Success of Some Methods for Measuring and Describing the Shape of Complex Objects, Systematic Biology, Vol. 47, No. 2, pp. 264-281, June 1998. |
| Analyze protrusions and indentations | Activates the analysis of convex sections (indentations) and concave sections (protrusions) along a contour. This analysis results in 12 features:
The analysis can be configured with additional configuration parameters:
|
| Calculate skeleton branch features | Activates extraction of various features from the skeleton of a region. This analysis results in 6 features:
|
| Calculate concavity information | Activates the extraction of two features based on a local concavity measure:
The analysis can be configured with additional configuration parameters:
For further details refer to H. Freeman, L.S. Davis, "A Corner-Finding Algorithm for Chain-Coded Curves," IEEE Transactions on Computers, vol. 26, no. 3, pp. 297-303, March 1977. |
| Calculate convex hull measures | Activates the extraction of some features based on the convex hull of a region:
The convex hull of a region is calculated using the Jarvis-March-Algorithm. |
In addition to these parameters for configuring which features to extract some more optional parameters are provided which do not have any influence on the feature extraction, i.e. do not change the results:
| Name of Option | Description |
| Fractional digits | allows to specify the accuracy in calculating features, i.e. the number of decimal places used in the result tables |
| Show curvature analysis info image? | if enabled, an additional result image is generated visualizing features based on curvature |
| Show skeleton analysis info image? | if enabled, an additional result image is generated visualizing skeleton features |
| Verbose | turns on output of additional debug messages; the messages are shown in the console |
Note that some of the features depend on others, e.g., extracting the circularity automatically results in extracting region area and perimeter as well.
Result data
The operator basically returns a data table. Each row in the table refers to one region, each column to a certain feature.
Note that the first column contains the index of the region as it is extracted from the label image or region set.
If additional result data, e.g. images for visualizing curvature or skeleton information, have been requested, these are also generated by the operator.