Applications/PaCeQuant
PaCeQuant Plugin

PaCeQuant is available since release version 1.8.6 of MiToBo.
In MiToBo version 2.0 released in May 2020 it got an update which added the largest empty circle as shape criterion to PaCeQuant's feature set.
The PaCeQuant plugin as part of MiToBo as well as the R script for the visualization of result data are published under the terms of GNU General Public License v3.0.

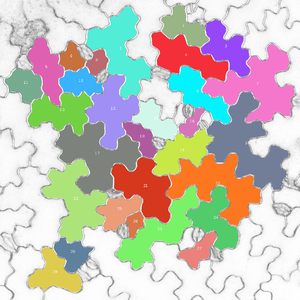
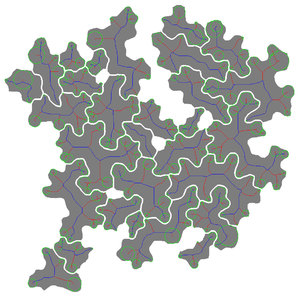
Name of Plugin/Operator
de.unihalle.informatik.MiToBo.apps.cellMorphology.PaCeQuant
(available since MiToBo version 1.8.6)
Related Publications
When using PaCeQuant, please cite us:
- Y. Poeschl, B. Möller, L. Müller, and K. Bürstenbinder,
User-friendly assessment of pavement cell shape features with PaCeQuant: Novel functions and tools.
In Methods in Cell Biology, Academic Press, 2020. - B. Möller, Y. Poeschl, S. Klemm, and K. Bürstenbinder,
Morphological Analysis of Leaf Epidermis Pavement Cells with PaCeQuant.
In F. Cvrčková and V. Žárský, editors, Plant Cell Morphogenesis: Methods and Protocols, chapter 22, pages 329-349, Springer New York, New York, NY, 2019. - B. Möller, Y. Poeschl, R. Plötner, and K. Bürstenbinder,
PaCeQuant: A Tool for High-Throughput Quantification of Pavement Cell Shape Characteristics.
Plant Physiology, 175(1), September 2017.
Main Features
- extraction of 28 characteristic shape features to quantify pavement cell shape
- fully automatic segmentation of cell regions from input images
- optional import of external/manual cell segmentation data
- classification of lobes into type I (2-cell contact) and type II (3-cell contact)
- additional R scripts for feature visualization
Supplemental Tools
The core PaCeQuant plugin has been supplemented by additional tools to ease feature visualization and analysis:
- the FeatureColorMapper plugin for creating heatmap visualizations of cell features
- the LabelImageEditor for handy manual post-processing of segmentation results
- the R package PaCeQuantAna for statistical analysis of feature data and creation of publication-ready plots of feature distributions and statistical analysis results
Usage - Parameters
To run PaCeQuant perform the following steps:
- install MiToBo by following the instructions on the Installation page
- run MiToBo and start the operator runner by selecting the menu item PaCeQuant from Plugins -> MiToBo
This will bring up the operator window of PaCeQuant.
Phases and Operation modes
PaCeQuant supports three different options for running listed below:
- SEGMENTATION_AND_FEATURES:
expects images as input, segments the images and extracts features - SEGMENTATION_ONLY:
expects images as input, just segments the cell regions, no feature extraction - FEATURES_ONLY:
works on binary or label images or ImageJ regions, extracts features for the given regions
In addition PaCeQuant can be run in either of two modes:
- INTERACTIVE:
PaCeQuant processes the data provided directly within the graphical environment of ImageJ, i.e. reads regions from the ROI manager, and directly shows results - BATCH:
PaCeQuant processes all files (images or ROI files) in a given folder and writes results to disk
For batch mode the user can specify an input directory, in interactive mode PaCeQuant expects an input image or regions to be available in ImageJ.
Depending on the chosen phase and operation mode the graphical user interface is dynamically re-configured to show only the options relevant for the selected options.
Input Image/Directory
Depending on the chosen phases and operation modes here you need to specify either input data already loaded in ImageJ/Fiji or a directory where PaCeQuant can find the data to process. In detail, you have to provide the following information for the various configurations:
| Operation Mode | Phase(s) to Run | Input Data |
| INTERACTIVE | SEGMENTATION_ONLY | Gray-scale input image already opened in ImageJ/Fiji. |
| SEGMENTATION_AND_FEATURES | Gray-scale input image already opened in ImageJ/Fiji. | |
| FEATURES_ONLY | Binary or label image already opened in ImageJ/Fiji, or ImageJ ROI set from ROI manager. | |
| BATCH | SEGMENTATION_ONLY | Directory containing gray-scale input images. First-level sub-folders are also processed. |
| SEGMENTATION_AND_FEATURES | Directory containing gray-scale input images. First-level sub-folders are also processed. | |
| FEATURES_ONLY | Directory containing either binary or label images to process, a collection of ImageJ ROI files (with ending '.roi'), or an archive of multiple ImageJ ROIs (with ending '.zip'). |
Important notice:
In BATCH mode PaCeQuant tries to analyze all image files present in the given folder or any of its direct sub-folders. In particular, PaCeQuant will also analyze data from a potential result folder of a previous run, if it is present in the given folder. To avoid problems resulting from analyzing wrong data you should ensure that the provided directory only contains native input images or segmentation data and no other image data not suitable for processing with PaCeQuant.
Calibration
As PaCeQuant measures lengths and areas from the given data it is important that the tool is properly calibrated, i.e. the physical size of a pixel is known. PaCeQuant supports two calibration modes:
- AUTO:
here PaCeQuant seeks to extract calibration information from the given input data - MANUAL:
the user can enter calibration data, i.e. the physical size of a pixel and the units to use
Important notice:
Please note that in ImageJ ROI files no calibration data is stored. Thus, when using PaCeQuant to extract features from external segmentation data provided as ImageJ ROIs, you always need to manually provide calibration data. And this also holds when manually post-processing segmentation data extracted with PaCeQuant as all calibration data of the original images will get lost.
Configuration of Segmentation Phase
For detailed configuration of the algorithms applied during the segmentation phase the following parameters are available:
| Name | Description |
| Border Contrast | Allows to select whether the boundaries of the cells are darker or brighter than the background. |
| Heuristic for Gap Closing | During segmentation sometimes small gaps in the boundaries remain which can be closed by PaCeQuant applying one of the following heuristics:
|
| Unit for Size Thresholds | Unit in which the size thresholds for filtering valid regions (see next two parameters) are specified, i.e. either PIXELS or MICRONS. |
| Minimal Size of Cells | Segmented cells being too small can automatically be excluded by specifying a minimal size for valid cells. |
| Maximal Size of Cells | Segmented cells being too large can also automatically be excluded by specifying a maximal size for valid cells. |
Configuration of Feature Extraction Phase
| Name | Description |
| Feature Extraction | For changing various parameters used in feature extraction the MorphologyAnalyzer2D operator of MiToBo is used and can be configured here. Note that changing parameters might hamper comparison of PaCeQuant results among different work groups or laboratories. |
| Analyze lobe types? | Activates the optional classification of individual lobes into type I (2-cell contact) or type II (3-cell contact) lobes. |
Additional Configuration Parameters
| Name | Description |
| Niblack threshold | Allows to configure the binarization step in the segmentation phase, i.e., to increase or decrease the sensitivity of PaCeQuant for cell boundaries. The lower the threshold the more potential boundary candidate pixels will be extracted which might add to an improved segmentation in case of low contrast. |
| Draw region IDs to output image? | If the segmentation phase is run PaCeQuant outputs a label image showing segmented cell regions. If this option is activated region IDs are drawn to that image for easier interpretation. But note that this renders the image unsuitable for any further automatic analysis. |
| Verbose | If enabled additional log messages will be printed to console. |
| Show/save additional results? | If enabled an additional image stack with a collection of intermediate images will be generated. The images in this stack might help to get a deeper insight into PaCeQuant and might help to identify problems in case that the segmentation of input images fails. |
| Show/save feature stack? | If enabled a stack of images is generated where each image visualizes the values of a specific feature. For most images in this stack the feature values of individual cells are mapped to the intensity value of the corresponding cell (e.g., for features like area, solidity, width, length, branch count, etc.). |
Usage - Output data
If PaCeQuant is run in INTERACTIVE mode all results are directly shown in ImageJ/Fiji, while in BATCH mode all result data is saved to a folder named results in the input folder. Which kinds of output data are resulting from running PaCeQuant depends on the selected phases:
- list of result files from SEGMENTATION phase:
- *-allRois.zip: ImageJ ROI archive containing all ROIs extracted from a single image
- *_<number>.roi: ROI file containing only ROI with ID numberextracted from single image
- *-grayscale-result.tif: gray-scale label image of extracted cell regions
- *-color-result.tif: overlay of extracted cell regions (in pseudo-color) over input image
- *-intermediate-result-stack.tif: (optional) stack of intermediate result images
- list of result files from FEATURES phase:
- *-allRois-table.txt: extracted feature values for all cells in a given image
- *-allRois-grayscale-result.tif: gray-scale label image showing analyzed regions
- *-intermediate-result-stack.tif: (optional) stack of intermediate result images
- *-feature-stack.tif: (optional) stack of images visualizing feature values for cells
- *-allRois-lobe-types.tif: (if lobe type classification is enabled) image visualizing the type of each lobe of all cells analyzed
- *-allRois-cell-<number>-lobe-table.txt: (if lobe type classification is enabled) file containing features for lobes of cell with ID number
Note that the asterisk in the above file names represents a single image, i.e. the set of files exists for each image present in the given folder.
All files with ending *.ald are internal MiToBo log files in XML format where, e.g., parameter settings are stored and which usually can be ignored or just preserved for later reference.
Sample data
Here we provide some sample data with which you can test your local PaCeQuant installation:
To apply PaCeQuant to the test data simply unpack the data archive to a folder of your choice and set the input directory parameter of PaCeQuant to the sample data folder.
The following archives contain result data extracted from the sample data with PaCeQuant and default settings for validating if your local PaCeQuant installation yields correct results.
Depending on the PaCeQuant version the result data slightly differs:
- Result data PaCeQuant v2.0 (May 2020): sampleDataResults-v2.0.zip (Java version: 1.8)
Starting with version 2.0 PaCeQuant provides the Largest Empty Circle feature, and in this version also an issue with measurement units in lobe measurings has been fixed.
- Result data PaCeQuant v1.0 (Sep 2017): sampleDataResults-v1.0.zip
Please note that we occasionally encountered marginal deviations in numerical feature measurements in the last decimals between different versions of Java.
In particular with regard to curvature based measures which rely on trigonometric functions small changes in the values below the sixth decimal were sometimes observed.
Complementary Tools: FeatureColorMapper, LabelImageEditor and PaCeQuantAna
Over the time several complementary tools for PaCeQuant have been developed which ease manual correction of segmentation results and simplify analysis and exploration of extracted shape features:
- the FeatureColorMapper plugin for creating heatmap visualizations of cell features
- the LabelImageEditor for handy manual post-processing of segmentation results
- the R package PaCeQuantAna for statistical analysis of feature data and creation of publication-ready plots of feature distributions and statistical analysis results
Notes on manual or semi-automatic segmentation of cells
PaCeQuant supports extracting features from externally provided segmentation data.
This can be helpful in cases where the original input images are not suitable for automatic segmentation by PaCeQuant,
e.g., due to a general low image quality or image acquisition techniques for which PaCeQuant is not optimized.
The segmentation data can be provided in different formats:
- an 8-bit binary image where all cell regions are labeled white (intensity = 255)
- an 8-bit gray-scale image where each cell region is marked with an individual label (intensity larger than zero) and the background in black (intensity = 0)
- ImageJ ROI files
Note that when providing binary images neighboring regions must not touch each other. Optimally the margins between neighboring cell regions will have a width of at least 3 pixels.
ImageJ ROI files
For providing segmentation data in terms of ImageJ ROI files the easiest way to do this is to segment regions directly in ImageJ/Fiji.
Use the Polygon Section Tool for segmenting individual cells, add each of the polygons to the ROI manager, and finally save all ROIs to a file.
In case that you aim to improve an initial segmentation result of PaCeQuant, open the ROI files generated by PaCeQuant during the segmentation phase with ImageJ's ROI manager and edit the ROIs.
For editing polygon selections ImageJ offers a large variety of possibilities, e.g.:
- single points can be removed (Ctrl + click)
- new points can be added by splitting segments (Shift + click)
- the complete region can be moved
- the polygon can be interpolated, i.e. sub-sampled
More information on the various options can be found in the ImageJ documentation:
Probably of interest might also be the macro 'Modifying Polygon Selections'.